Live
-
{{ is_pkg(link.U) }}{{ link.T }} {{ get_host(link.U) }} - {{ link.N }} ( {{ get_host(link.U) }} )
{{ is_pkg(link.U) }}{{ link.T }} {{ get_host(link.U) }}( {{ get_host(link.U) }} )
{{ item.date }}
-
{{ is_pkg(link.U) }}{{ link.T }} {{ get_host(link.U) }} - {{ link.N }}
( {{ get_host(link.U) }} ){{ is_pkg(link.U) }}{{ link.T }} {{ get_host(link.U) }}
( {{ get_host(link.U) }} )
R Weekly 2026-W07 sitrep, webRios, Jarl linter
This week’s release was curated by Eric Nantz, with help from the R Weekly team members and contributors.
Highlight
Insights
-
Paleontology R Packages to Benefit from Software Sustainability Institute Grant
-
Announcing New Statistical Software Peer Review Editors: Natalia da Silva and Andrew Heiss
-
Why Every R Package Wrapping External Tools Needs a sitrep() Function
-
Announcing the Positron Notebook Editor for Jupyter Notebooks
-
Fifty Releases of Pointblank: a Year of Building Data Quality Tooling
R in the Real World
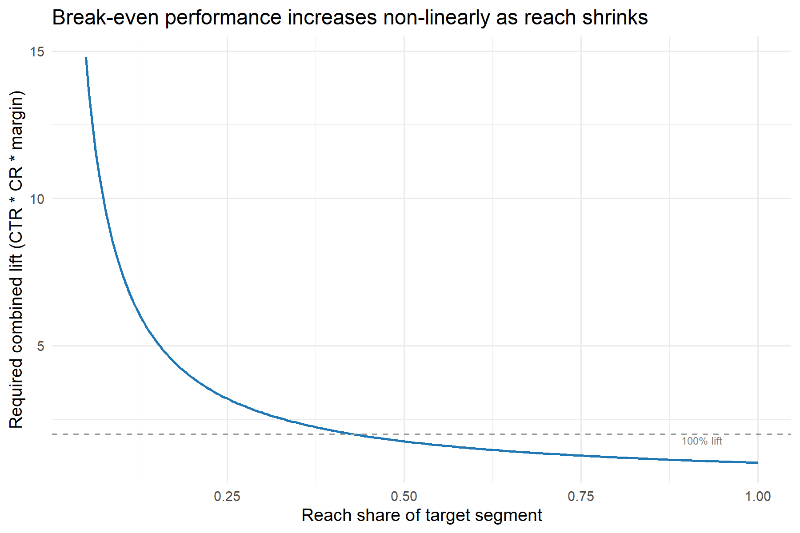
R in Organizations
R in Academia
Tutorials
Resources
New Packages
📦 Keep up to date wtih CRANberries 📦
CRAN
- {whisper} 0.1.0: Native R ‘torch’ Implementation of ‘OpenAI’ ‘Whisper’
- {vascr} 0.1.4: Process Biological Impedance Sensing Data
- {tirt} 0.1.3: Testlet Item Response Theory
- {nlmixr2auto} 1.0.0: Automated Population Pharmacokinetic Modeling
- {keyed} 0.1.3: Explicit Key Assumptions for Flat-File Data
- {indonesiaFootballScoutR} 0.1.3: Tools for Football Player Scouting in Indonesia
- {bioLeak} 0.1.0: Leakage-Safe Modeling and Auditing for Genomic and Clinical Data
- {bayesics} 2.0.2: Bayesian Analyses for One- and Two-Sample Inference and Regression Methods
- {BayesianHybridDesign} 0.1.0: Bayesian Hybrid Design and Analysis
- {areaOfEffect} 0.2.4: Spatial Support at Scale
- {wired} 1.0.0: Weighted Adaptive Prediction with Structured Dependence
- {snreg} 1.2.0: Regression with Skew-Normally Distributed Error Term
- {resLIK} 0.1.2: Representation-Level Control Surfaces for Reliability Sensing
- {PPtreeExt} 0.1.0: Projection Pursuit Classification Tree Extensions
- {mortSOA} 0.1.0: Obtain Data from the Society of Actuaries ‘Mortality and Other Rate Tables’ Site
- {tidylearn} 0.1.0: A Unified Tidy Interface to R’s Machine Learning Ecosystem
- {quickSentiment} 0.1.0: A Fast and Flexible Pipeline for Text Classification
- {numspellR} 0.1.0: Detection of Numeric Persistence and Rigidity Patterns
- {climenu} 0.1.3: Interactive Command-Line Menus
- {roxigraph} 0.1.0: ‘RDF’ and ‘SPARQL’ for R using ‘Oxigraph’
- {regextable} 0.1.1: Pattern-Based Text Extraction and Standardization with Lookup Tables
- {wlsd} 1.0.1: Wrangling Longitudinal Survival Data
- {Orangutan} 2.0.0: Automated Analysis of Phenotypic Data
- {marinepredator} 0.0.1: Marine Predators Algorithm
- {GalaxyR} 0.1.0: ‘Galaxy’ API Implementation
- {directadjusting} 0.6.1: Directly Adjusted Estimates
- {SNMA} 0.1.5: Stream Network Movement Analyses
- {ppweibull} 1.0: Piecewise Lifetime Models
- {gsDesignTune} 0.1.0: Dependency-Aware Scenario Exploration for Group Sequential Designs
- {avstrat} 0.1.1: Stratigraphic Data Processing and Section Plots
- {typeR} 0.2.0: Simulate Typing Script
- {tidyextreme} 1.0.0: A Tidy Toolbox for Climate Extreme Indices
- {sqlm} 0.1.0: SQL-Backed Linear Regression
- {setweaver} 1.0.0: Building Sets of Variables in a Probabilistic Framework
- {CurricularComplexityData} 0.1.0: Data for Exploring Curricular Complexity
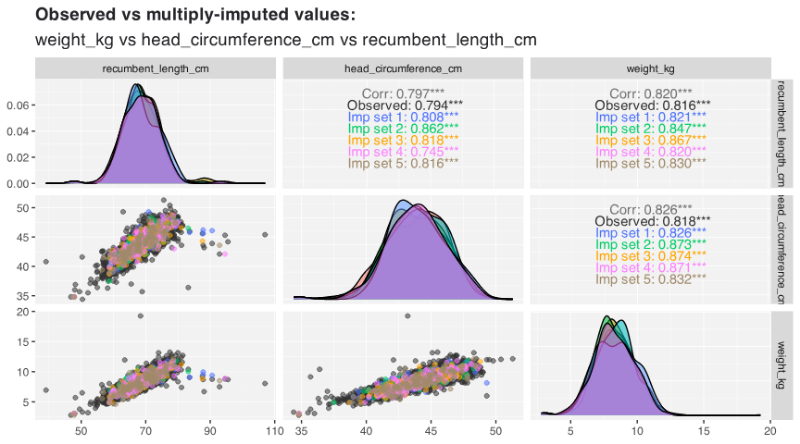
- {vismi} 0.9.5: Visual Diagnostics for Multiple Imputation
- {sleacr} 0.1.3: Simplified Lot Quality Assurance Sampling Evaluation of Access and Coverage (SLEAC) Tools
- {rcatfish} 1.0.0: An R Interface to the California Academy of Sciences Eschmeyer’s Catalog of Fishes
- {SepTest} 0.0.1: Tests for First-Order Separability in Spatio-Temporal Point Processes
- {SemiparMF} 1.0.0: Semiparametric Spatiotemporal Model with Mixed Frequencies
- {scenfire} 0.1.0: Post-Processing Algorithm for Integrating Wildfire Simulations
- {multiRec} 1.0.6: Analysis of Multi-Type Recurrent Events
- {gipsDA} 0.1.2: Training DA Models Utilizing ‘gips’
- {findn} 0.1.0: Simulation Based Sample Size Estimation
- {educabR} 0.1.0: Download and Process Brazilian Education Data from INEP
- {sasif} 0.1.2: ‘SAS’ IF Style Data Step Logic for Data Tables
- {phinterval} 1.0.0: Set Operations on Time Intervals
- {multiSA} 0.1.0: Multi-Stock Assessment
- {MADtests} 0.1.1: Hypothesis Tests and Confidence Intervals for Median Absolute Deviations
- {baselinenowcast} 0.2.0: Baseline Nowcasting for Right-Truncated Epidemiological Data
- {spanner} 1.0.2: Utilities to Support Lidar Applications at the Landscape, Forest, and Tree Scale
- {opusreader2} 0.6.8: Read Spectroscopic Data from Bruker OPUS Binary Files
- {nomiShape} 1.0.0: Visualization and Analysis of Nominal Variable Distributions
- {healthbR} 0.1.1: Access Brazilian Public Health Data
- {CGMissingDataR} 0.0.1: Missingness Benchmark for Continuous Glucose Monitoring Data
- {rotulador} 1.0.0: Useful Functions for Programming and Generating Documents
- {fluxfixer} 1.0.0: Advanced Framework for Sap Flow Data Post-Process
- {blindspiker} 0.2.0: Laboratory Blind Spike Sample Analyses
- {BioGSP} 1.0.0: Biological Graph Signal Processing for Spatial Data Analysis
- {SVG} 1.0.0: Spatially Variable Genes Detection Methods for Spatial Transcriptomics
- {date4ts} 0.1.1: Wrangle and Modify Ts Object with Classic Frequencies and Exact Dates
- {comphy} 1.0.5: Functions Used in the Book “Computational Physics with R”
- {blockstrap} 1.0.0: Sample Dataframes by a Group
- {adsasi} 0.9.0.1: Adaptive Sample Size Simulator
GitHub or Bitbucket
- {timenow}: Quick Timezone-aware Timestamps
Updated Packages
- {vazul} 1.1.0: Analysis Blinding Tools - diffify
- {scorecard} 0.4.6: Credit Risk Scorecard - diffify
- {reporter} 1.4.6: Creates Statistical Reports - diffify
- {partykit} 1.2-25: A Toolkit for Recursive Partytioning - diffify
- {mlpack} 4.7.0: ‘Rcpp’ Integration for the ‘mlpack’ Library - diffify
- {gratia} 0.11.2: Graceful ‘ggplot’-Based Graphics and Other Functions for GAMs Fitted Using ‘mgcv’ - diffify
- {bibliorefer} 0.1.3: Generator of Main Scientific References - diffify
- {bcdata} 0.5.2: Search and Retrieve Data from the BC Data Catalogue - diffify
- {osrm.backend} 0.2.0: Bindings for ‘Open Source Routing Machine’ - diffify
- {crosswalkr} 0.4.0: Rename and Encode Data Frames Using External Crosswalk Files - diffify
- {UCSCXenaTools} 1.7.0: Download and Explore Datasets from UCSC Xena Data Hubs - diffify
- {SomaDataIO} 6.5.0: Input/Output ‘SomaScan’ Data - diffify
- {shinyscholar} 0.4.4: A Template for Creating Reproducible ‘shiny’ Applications - diffify
- {OmopSketch} 1.0.1: Characterise Tables of an OMOP Common Data Model Instance - diffify
- {feisr} 1.3.1: Estimating Fixed Effects Individual Slope Models - diffify
- {eurlex} 0.4.9: Retrieve Data on European Union Law - diffify
- {BioCro} 3.3.1: Modular Crop Growth Simulations - diffify
- {profileModel} 0.6.2: Profiling Inference Functions for Various Model Classes - diffify
- {ipeaplot} 0.5.1: Add Ipea Editorial Standards to ‘ggplot2’ Graphics - diffify
- {gooseR} 0.1.2: R Integration for ‘Goose’ AI - diffify
- {vigicaen} 1.0.0: ‘VigiBase’ Pharmacovigilance Database Toolbox - diffify
- {micar} 1.2.0: ‘Mica’ Data Web Portal Client - diffify
- {AutoPlots} 1.5.0: Creating Echarts Visualizations as Easy as Possible - diffify
- {tidydfidx} 0.0-3: Indexed ‘tibble’ and Methods for ‘dplyr’ - diffify
- {resourcecode} 0.5.3: Access to the ‘RESOURCECODE’ Hindcast Database - diffify
- {gifti} 0.9.0: Reads in ‘Neuroimaging’ ‘GIFTI’ Files with Geometry Information - diffify
- {resourcer} 1.5.0: Resource Resolver - diffify
- {SBMTrees} 1.4: Longitudinal Sequential Imputation and Prediction with Bayesian Trees Mixed-Effects Models for Longitudinal Data - diffify
- {permute} 0.9-10: Functions for Generating Restricted Permutations of Data - diffify
- {bindrcpp} 0.2.4: An ‘Rcpp’ Interface to Active Bindings - diffify
- {simPH} 1.3.15: Simulate and Plot Estimates from Cox Proportional Hazards Models - diffify
- {Ruido} 1.0.2: Soundscape Background Noise, Power, and Saturation - diffify
- {qryflow} 0.2.0: Execute Multi-Step ‘SQL’ Workflows - diffify
- {traumar} 1.2.4: Calculate Metrics for Trauma System Performance - diffify
- {tidychangepoint} 1.0.4: A Tidy Framework for Changepoint Detection Analysis - diffify
- {Rparadox} 0.2.0: Read Paradox Database Files into R - diffify
- {idmc} 0.4.1: Load and Wrangle IDMC Displacement Data - diffify
- {shinyloadtest} 1.2.1: Load Test Shiny Applications - diffify
- {phsopendata} 1.0.3: Extract from the Scottish Health and Social Care Open Data Platform - diffify
- {penetrance} 0.1.2: Methods for Penetrance Estimation in Family-Based Studies - diffify
- {ISAR} 1.0.2: Introduction to Sports Analytics using R (ISAR) Data - diffify
- {HVT} 26.1.2: Constructing Hierarchical Voronoi Tessellations and Overlay Heatmaps for Data Analysis - diffify
- {tidyCDISC} 0.2.2: Quick Table Generation & Exploratory Analyses on ADaM-Ish Datasets - diffify
- {BFS} 0.7.1: Get Data from the Swiss Federal Statistical Office - diffify
- {fru} 0.0.3: A Blazing Fast Implementation of Random Forest - diffify
- {doRNG} 1.8.6.3: Generic Reproducible Parallel Backend for ‘foreach’ Loops - diffify
- {statsExpressions} 1.7.3: Tidy Dataframes and Expressions with Statistical Details - diffify
- {aion} 1.7.0: Archaeological Time Series - diffify
- {cardargus} 0.2.1: Generate SVG Information Cards with Embedded Fonts and Badges - diffify
- {discovr} 1.0.0: Interactive Tutorials and Data for “Discovering Statistics Using R and RStudio” - diffify
- {chunked} 0.6.2: Chunkwise Text-File Processing for ‘dplyr’ - diffify
- {pmparser} 1.0.24: Create and Maintain a Relational Database of Data from PubMed/MEDLINE - diffify
- {efdm} 0.2.2: Simulate Forest Resources with the European Forestry Dynamics Model - diffify
- {COMIX} 1.0.2: Coarsened Mixtures of Hierarchical Skew Kernels - diffify
- {collections} 0.3.11: High Performance Container Data Types - diffify
- {cardx} 0.3.2: Extra Analysis Results Data Utilities - diffify
- {PAMscapes} 0.15.0: Tools for Summarising and Analysing Soundscape Data - diffify
- {vowels} 1.2-3: Vowel Manipulation, Normalization, and Plotting - diffify
- {kDGLM} 1.2.12: Bayesian Analysis of Dynamic Generalized Linear Models - diffify
- {kindling} 0.2.0: Higher-Level Interface of ‘torch’ Package to Auto-Train Neural Networks - diffify
- {BayesMallowsSMC2} 0.2.1: Nested Sequential Monte Carlo for the Bayesian Mallows Model - diffify
- {PaRe} 0.1.16: A Way to Perform Code Review or QA on Other Packages - diffify
- {goat} 1.1.5: Gene Set Analysis Using the Gene Set Ordinal Association Test - diffify
- {ediblecity} 0.2.2: Modeling Urban Agriculture at City Scale - diffify
- {restatapi} 0.24.5: Search and Retrieve Data from Eurostat Database - diffify
- {tidywikidatar} 0.6.0: Explore ‘Wikidata’ Through Tidy Data Frames - diffify
- {insight} 1.4.6: Easy Access to Model Information for Various Model Objects - diffify
- {fanyi} 0.1.1: Translate Words or Sentences via Online Translators - diffify
- {lubridate} 1.9.5: Make Dealing with Dates a Little Easier - diffify
- {beast} 1.2: Bayesian Estimation of Change-Points in the Slope of Multivariate Time-Series - diffify
- {viridisLite} 0.4.3: Colorblind-Friendly Color Maps (Lite Version) - diffify
- {wizaRdry} 0.6.4: A Magical Framework for Collaborative & Reproducible Data Analysis - diffify
- {bmemLavaan} 0.7: Mediation Analysis with Missing Data and Non-Normal Data - diffify
- {layer} 0.0.4: Tilt your Maps and Turn Them into ‘ggplot’ Plots - diffify
Videos and Podcasts
R Project Updates
Updates from R Core:
Upcoming Events in 3 Months
Events in 3 Months:
-
A gRadual Intro to Shiny - R-Ladies Remote (February 17, 2026)
Connect
Join the Data Science Learning Community